Device to combat memory loss from brain injury, epilepsy, Alzheimer’s disease created
Posted by Acubiz | BlogUT Southwestern Medical Center has joined a consortium of seven leading universities to develop new technologies to improve memory in people with traumatic brain injury, mild cognitive impairment, epilepsy, and Alzheimer’s disease.
Specifically, UT Southwestern is part of a study with the goal of developing an implantable neural monitoring and stimulation system by the end of 2018 that would treat memory loss.
Researchers plan to use safe levels of electrical stimulation to test new ways of improving brain function and memory in neurosurgery patients who already receive brain stimulation as part of their therapy for epilepsy. Their goal is to determine whether brain stimulation delivered when these individuals play memory games will improve their memory ability.
“If memory can be improved in patients who have electrodes implanted to treat epilepsy − and who frequently have mild memory impairment − then we will have gained valuable information on how to restore memory function in patients with traumatic brain injury or Alzheimer’s disease,” said Dr. Bradley Lega, Assistant Professor of Neurological Surgery, Neurology and Neurotherapeutics, and Psychiatry, who leads the Dallas arm of the study.
The effort is part of a national “Restoring Active Memory” (RAM) program sponsored by the Defense Advanced Research Projects Agency and supported by the National Institutes of Health’s Brain Research through Advancing Innovative Neurotechnologies (BRAIN). To date, the NIH and other funding sources have allocated more than $240 million to the BRAIN Initiative. The initiative is designed to improve understanding of the brain and cognitive function by accelerating the development and application of innovative technologies to find new ways to treat, cure, and even prevent brain disorders.
In 2015, an estimated 5.3 million Americans had Alzheimer’s disease, according to the Alzheimer’s Association, while about 2.9 million Americans currently have epilepsy, according to Centers for Disease Control and Prevention (CDC) estimates. The CDC also estimates 1.7 million traumatic brain injuries occur annually in the U.S.
Dr. Lega is recruiting 15 patients per year with epilepsy to undergo stereo electroencephalography (sEEG), a minimally invasive technique for recording brain waves to diagnose epilepsy. Dr. Lega is one of the few neurosurgeons in the country who uses stereo EEG to locate the origin of epileptic seizures in the brain and to determine if a patient is a candidate for surgery to treat the seizures. Less invasive than the traditional approach, stereo EEG involves electrodes placed in the brain to record electrical activity during seizures.
The data gathered at UT Southwestern will be combined with data from the University of Pennsylvania, Thomas Jefferson University in Philadelphia, the Mayo Clinic, Dartmouth University, Emory University, and Boston University to develop and test new treatments.
“The national research team believes that the therapeutic strategies being examined in this study will serve as the foundation for novel brain-machine interface devices that will improve memory function,” Dr. Lega said.
Next-generation immunotherapy offers new hope for beating brain cancer
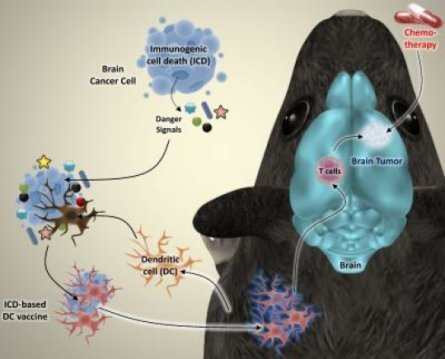
Posted by Acubiz | BlogThe researchers induced a specific type of cell death in brain cancer cells from mice. The dying cancer cells were then incubated together with dendritic cells, which play a vital role in the immune system. The researchers discovered that this type of cancer cell killing releases ‘danger signals’ that fully activate the dendritic cells. “We re-injected the activated dendritic cells into the mice as a therapeutic vaccine”, Professor Patrizia Agostinis explains. “That vaccine alerted the immune system to the presence of dangerous cancer cells in the body. As a result, the immune system could recognize them and start attacking the brain tumor.”
Credit: ©KU Leuven Laboratory of Cell Death Research & Therapy – Dr. Abhishek D. Garg
High-grade glioma is the most aggressive form of brain cancer. Despite improvements in surgical procedures, chemotherapy, and radiotherapy, this type of brain tumour is still notoriously hard to treat: less than 10% of patients survive beyond five years. Researchers from KU Leuven, Belgium, have now shown that next-generation cell-based immunotherapy may offer new hope in the fight against brain cancer.
Cell-based immunotherapy involves the injection of a therapeutic anticancer vaccine that stimulates the patient’s immune system to attack the tumour. Thus far, the results of this type of immunotherapy have been mildly promising. However, Abhishek D. Garg and Professor Patrizia Agostinis from the KU Leuven Department of Cellular and Molecular Medicine have now found a novel way to produce more effective cell-based anticancer vaccines.
The researchers induced a specific type of cell death in brain cancer cells from mice. The dying cancer cells were then incubated together with dendritic cells, which play a vital role in the immune system. The researchers discovered that this type of cancer cell killing releases ‘danger signals’ that fully activate the dendritic cells.
“We re-injected the activated dendritic cells into the mice as a therapeutic vaccine,” Professor Patrizia Agostinis explains. “That vaccine alerted the immune system to the presence of dangerous cancer cells in the body. As a result, the immune system could recognize them and start attacking the brain tumour.”
Combined with chemotherapy, this novel cell-based immunotherapy drastically increased the survival rates of mice afflicted with brain tumours. Almost 50% of the mice were completely cured. For the sake of comparison: none of the mice treated with chemotherapy alone became long-term survivors.
“The major goal of any anticancer treatment is to kill all cancer cells and prevent any remaining malignant cells from growing or spreading again,” Professor Agostinis continues. “This goal, however, is rarely achieved with current chemotherapies, and many patients relapse. That’s why the co-stimulation of the immune system is so important for cancer treatments. Scientists have to look for ways to kill cancer cells in a manner that stimulates the immune system. With an eye on clinical studies, our findings offer a feasible way to improve the production of vaccines against brain tumours.”
A human liver microphysiology platform for studying physiology, drug safety, and disease
Posted by Acubiz | BlogThe human body critically depends on the liver to metabolize toxins and synthesize biomolecules necessary for life. In addition to being a site for life threatening diseases, the liver is particularly sensitive to damage induced by xenobiotics and drugs. Lawrence Vernetti, a co-author, puts it this way: “The cost to the drug developer becomes enormous if unexpected liver damage emerges late in drug development. You either stop development of potentially life-saving drugs or face additional expensive human clinical trials.”
Liver toxicity and disease is also influenced by liver resident cell types other than hepatocytes and by the microenvironment, such as regions where hepatocytes are known to exist in reduced oxygen conditions. While there are several in vitro human liver models, none of the models provides for long-term chronic exposure studies, while mimicking the physiological conditions created by immune, stellate and endothelial cells, and supporting the detailed detection of early indications of cellular dysfunction by a combination of direct imaging and biochemical readouts.
A study by D. Lansing Taylor and colleagues in the January 2016 Issue (241:1) of Experimental Biology and Medicine reports on the development and application of the first generation, Sequentially Layered, Self-Assembly Liver (SQL-SAL) model, a four cell, human organ model constructed in an optically transparent microfluidic chamber. A key feature of the SQL-SAL model is the inclusion of fluorescent protein biosensors which are used for real-time monitoring of cellular functions such as apoptosis or generation of reactive oxygen species. To establish the predictivity of the model, a Microphysiology Systems (MPS) database that combines the experimental model data with drug-organ interaction data accessed from a variety of public and private databases has been created.
Performance of the SQL-SAL over 28 days was demonstrated by time and dose-dependent changes in liver functions and activation of key toxicity pathways in response to clinically relevant hepatotoxins. Furthermore, hepatotoxins produced immune-cell mediated hepatocellular damage or the deposition of collagen linked to fibrosis concordant with known clinical findings. As D. Lansing Taylor, Director of the University of Pittsburgh Drug Development Institute and the Principle Investigator for the SQL-SAL explains, “The results so far, demonstrate that the 3D, microfluidic, human liver model offers the drug research community a unique platform to test drugs at reasonable costs, early in the development cycle. With further development the SQL-SAL will be constructed using stem cell derived hepatocytes and other cell types from diseased and normal population to produce a powerful new tool to study disease progression and effectiveness of drugs.”
Diabetic management: Subcutaneous insulin therapy fails to protect against oxidative stress and inflammation
Posted by Acubiz | BlogSubcutaneous insulin infusion (CSII) is the gold standard for type 1 diabetic patient therapy. Less physiological than intraperitoneal administration, the subcutaneous route may induce glycemic variability in some patients, a powerful enhancer of reactive oxygen species (ROS) production. While this oxidative stress is recognized to play a role in diabetes and its complications, its characterization has not been fully achieved, especially in the liver, the target organ for insulin sensitivity. Under physiological conditions, an endogenous antioxidant system ensures the oxidative balance. Host survival depends upon the ability of cells and tissues to adapt to or resist the stress and repair or remove damaged molecules and cells.
Sigrist and colleagues at the European Center of Diabetes Study (CEED, Strasbourg, France) have reported in the January 2016 issue ofExperimental Biology and Medicine that, in a diabetic rat model, a rapid increase of hepatic oxidative stress and inflammation biomarkers were observed, which is associated with drastic decrease of glycogen storage and protein synthesis. Continuous administration of insulin subcutaneously, using an osmotic mini-pump (CSII), rapidly decreased oxidative stress in liver and plasma, but failed after longer diabetes status. Moreover, hepatic and systemic inflammation was not prevented and a high variability of glycogen content was observed. In fact, CSII was not able to preserve the balance of anti- and pro-oxidant species. “Favoring a more physiological pathway for insulin administration would be a real advantage for better glycemic control, preserving organs from glucotoxicity-induced disorders and oxidative stress” stated Stéphanie DAL. These data support, for the first time, that targeting oxidative stress and inflammation could be a new therapeutic approach since conventional insulin therapy does not allow protection of the liver from chronic diabetes effects.
Cell biology: Nuclear export of opioid growth factor receptor is CRM1 dependent
Posted by Acubiz | BlogIn a study in the February 2016 Issue (241:3) ofExperimental Biology and Medicine researchers at The Pennsylvania State University College of Medicine, led by Dr. Pat McLaughlin, discovered that a novel biological pathway, the OGF-OGFr axis, regulates cell proliferation in normal and abnormal cells and tissues. The non-classical opioid receptor, OGFr, was first discovered in the 1980s in neural cancer cells and normal rodent brain through collaborative work with Drs. Ian S. Zagon and Steven R. Goodman, and subsequently was isolated, characterized, and cloned. The molecular and protein structure of OGFr has no resemblance to classical opioid receptors, and is an intrinsically unstructured protein with approximately 78% amino acid identity between mouse, rat, and human. OGFr gene and protein expression has been recorded in cells and tissues arising from all 3 dermal derivatives, and OGFr binding has been measured in a wide variety of developing and adult tissues, as well as more than 30 human cancer cell lines.
Dysregulation of the OGFr has been documented in several human cancers whereby biopsies of advanced-stage tumors have reduced receptor relative to normal or non-malignant tissues. The loss of OGFr limits the effectiveness of its specific inhibitory growth factor ligand, OGF, and the blockade of receptor function with low dosages of antagonists such as naltrexone. Thus intact OGFr is requisite for the OGF-OGFr pathway to mediate cell replication.
Blockade of OGFr by antagonists such as naltrexone has led to a variety of therapeutic interventions. The duration of blockade has impacted many biological pathways. Short-term blockade of OGFr with low dosages of naltrexone is currently being used for treatment of a variety of autoimmune diseases. The impact of this opioid antagonist biotherapy is broad-based. Approximately 52 million individuals in the US may benefit from either low dosages of naltrexone or complete receptor blockade for treatment of cancer, multiple sclerosis, inflammatory bowel disorders, or complications associated with diabetes. Worldwide, the potential audience that could benefit from biotherapeutics related to modulation of the OGF-OGFr axis approaches more than 350 million. Knowledge of the mechanism and pathways of OGFr can be used as a diagnostic for dysregulation of the OGF-OGFr regulatory pathway in a variety of diseases. It is anticipated that drug discovery researchers will identify specific and selective receptor antagonists that can be used as safe, inexpensive treatment of these disorders.
OGFr is required to translocate into the nucleus to facilitate its role in cell cycle regulation, and does so utilizing nuclear localization signals, and β and Ran proteins. However, the mechanism of OGFr export was unknown. The discovery, reported in the February 2016 issue of Experimental Biology and Medicine, provides the first evidence that OGFr export is CRM1 (chromosome region maintenance 1; aka exportin1 or Xpo1) dependent. The present study demonstrated in COS-7 (primate fibroblast-like) cells that endogenous, as well as exogenous OGFr accumulates in the nucleus following treatment with leptomycin B, indicating that OGFr is exported in a CRM1 dependent manner. The OGFr sequence contains one predicted nuclear export signal at residues 217-225. Examination of poly-ubiquination sites ruled out OGFr degradation in the nucleus and confirmed proteasome degradation of the receptor in the cytoplasm. Dimerization of OGFr also was not involved in export.
As Dr. Nancy Kren, who conducted the research for her doctoral thesis and is continuing postdoctoral study at the University of North Carolina explains, “What is particularly exciting about this is that the inhibitory effects of the OGF-OGFr axis are also dependent on an intact nuclear export signal. Moreover, the inhibitory function of OGFr appears to require 7 tandem repeats found in the c-terminus that are unique to the OGFr protein.” The novelty of these tandem repeats may lead to the identification of novel nuclear export pathways, and may also open new avenues of research for how OGFr is dysregulated in human disease.
As Dr. McLaughlin, Professor at Penn State College of Medicine, and senior author on this research and thesis advisor to Nancy Kren, suggests, “Our laboratory has extended our 30-year study of OGFr to now understand the mechanism of nuclear export. We are particularly interested in future work on these pathways in order to understand the role of the OGF-OGFr axis in a variety of diseases including cancer, autoimmune dysfunction, and complications to diabetes.”
Cancer patients with limited finances are more likely to have increased symptoms and poorer quality of life
Posted by Acubiz | BlogIf you’re a lung or colorectal cancer patient, what’s in your wallet could determine your level of suffering and quality of life during treatment, according to a new study by Dana Farber Cancer Institute researchers. The findings appear today in the Journal of Clinical Oncology.
“Most of the studies looking at financial stress look at what cancer does to your finances after diagnosis,” said Christopher Lathan, MD, MS, MPH, lead author of the study and a thoracic oncologist at Dana-Farber. “We were interested in looking at what happens when you have financial distress, defined in our study as little or no savings at the time of your diagnosis, and how that factor can impact quality of life.”
In the study, researchers looked to measure the association between patient financial strain and symptom burden and quality of life (QOL) for patients with new diagnoses of lung or colorectal cancer.
Patients participating in the Cancer Care Outcomes Research and Surveillance study were interviewed about their financial reserves, QOL, and symptom burden at 4 months of diagnosis and, for survivors, at 12 months of diagnosis. Researchers assessed the association of patient-reported financial reserves with patient-reported outcomes, including the Brief Pain Inventory, symptom burden on the basis of the European Organization for Research and Treatment of Cancer Quality of Life Questionnaire C30, and QOL on the basis of the EuroQoL-5 Dimension scale.
Among patients with lung and colorectal cancer, 40 percent and 33 percent, respectively, reported limited financial reserves. Relative to patients with more than 12 months of financial reserves, those with limited financial reserves reported significantly increased pain for lung and colorectal symptoms, greater symptom burden, and poorer QOL. With decreasing financial reserves, a clear dose-response relationship was present across all measures of well-being. These associations were also manifest for survivors reporting outcomes again at 1 year and persisted after adjustment for stage, co-morbidity, insurance, and other clinical attributes.
“We found that patients who had financial distress at the time of diagnosis were more likely to have poorer outcomes in physical and mental quality of life measures, pain, and symptom burden,” noted Lathan. “This effect persisted after adjusting for stage of disease, co-morbidity, income, age, and insurance.”
Researchers say the findings highlight the need to evaluate patients for potential financial distress at the time of diagnosis and to allow for clinicians to target appropriate resources and treatments for those patients who were already struggling before their cancer diagnosis.
A new way to discover DNA modifications
Posted by Acubiz | BlogDNA is made from four nucleosides, each known by its own letter — A, G, C, and T. However, since the structure of DNA was deciphered in 1953, scientists have discovered several other variants that are often added to the DNA sequences to replace one of the usual four letters.
These variants, which may be modified versions of the traditional nucleosides, often help cells to control which genes are turned on and off, and are referred to as “epigenetic marks” in the DNA. In bacteria, they can also protect DNA from invasion by other organisms such as viruses.
Until now, these DNA modifications have been discovered by chance, as scientists uncovered unexpected signals in chemical analyses of DNA. However, a new approach from MIT, the University of Florida, and other institutions offers a systematic approach to discovering unknown epigenetic marks and modifications.
“It’s a way to discover nucleic acid modifications that you didn’t know existed,” says Peter Dedon, the Underwood-Prescott Professor of Biological Engineering at MIT. “We’ve developed a technology platform for the discovery and characterization of these new modifications.”
Dedon and his colleagues suspect that bacteria and viruses, in particular, have many DNA modifications that have not been discovered yet, which could offer new antibiotic targets and new tools for biotechnology. Using their approach, which combines bioanalytical chemistry, comparative genomics, and a special type of DNA sequencing, the team has discovered a DNA modification that helps bacteria to protect their genomes from viral infection. They report the findings in the Proceedings of the National Academy of Sciences the week of Feb. 29.
A multipronged approach
DNA modifications are usually inserted by enzymes into DNA after it is synthesized during cell division. Such modifications are often used as markers that help tell a cell which genes should be turned on at a given time. The DNA modifications can also help cells defend their DNA from attack by foreign DNA from viruses and other bacteria. A larger variety of similar modifications also help all types of RNA, including messenger RNA and transfer RNA, perform their functions.
Dedon and Valérie de Crécy-Lagard, a professor of microbiology and cell science at the University of Florida, decided to take a more systematic approach to discovering such modifications.
De Crécy-Lagard had previously discovered many of the genes required for the synthesis of RNA modifications known as queuosine and archeosine, which are found in microorganisms and are derived from a common precursor called preQ0. Using comparative genomics, a technique for screening the genomes of different organisms for variants of specific DNA sequences, she found similar genes in multiple bacterial species, located in a specific gene cluster that contained genes for DNA modification. Comparative genomics, one leg of the platform, thus provided the first clue about a potential new DNA modification.
De Crécy-Lagard and Dedon, both senior authors of the new PNAS paper, decided to test de Crécy-Lagard’s prediction that these bacteria insert preQ0 into DNA. Using mass spectrometry, which allows researchers to both search for unknown molecules and find those with a specific mass, Dedon’s lab found DNA modified with preQ0-like structures, which the team named dADG, in bacteria with the modification gene cluster but not in bacteria that lacked it.
The researchers then showed that, in the types of bacteria they studied, the dADG modification is part of a defense system that protects the bacterial cell DNA. These bacteria produce enzymes called restriction enzymes that chop up the unmodified DNA of an invading virus, for example. Dedon and de Crécy-Lagard are now searching in other bacteria for different epigenetic functions for these DNA modifications.
In a collaboration with Richard Roberts, chief scientific officer at New England Biolabs, the team is now applying the third element of their technology platform to characterize the new dADG modification — a special kind of DNA sequencing called single-molecule real-time sequencing (SMRT), done on machines made by Pacific Biosciences. SMRT sequencing often picks up a signal when it encounters something other than the traditional four nucleosides.
“There’s a little hiccup in the process, a tiny little pause, and if you tune the software correctly, you can pick up that signal and know where it is in the genome,” Dedon says. “This lets you map DNA modifications across the genome.” This type of sequencing doesn’t reveal what the modification is, but it pinpoints its location.
By combining the observational power of SMRT sequencing with the predictive power of comparative genomics and the ability to detect and identify modifications using bioanalytics, scientists can find a new modification, figure out what the modification is, and discover its role.
Antibiotic targets
In humans, there are only a few known DNA modifications, most of which weren’t discovered until decades after the four traditional DNA bases were identified. “Are there more out there? It’s an interesting question,” Dedon says. “There probably aren’t a huge number, but we could go look for more in humans.”
In bacteria, on the other hand, the team believes there may be at least a dozen more modifications that haven’t been found yet, and even more in bacteriophages (viruses that infect bacteria). One of the important connections between bacterial DNA modifications and humans lies in the bacteria of the human gut — the gut microbiome. The team now has evidence that bacteria in the gut microbiome contain dADG, as well as another bacterial DNA modification that they discovered called phosphorothioates, and they are looking to see if they play any role in human health and disease.
Any new modifications discovered could also become useful antibiotic targets, especially those that prevent restriction enzymes from chopping up the bacteria’s own DNA. Drugs that inhibit the enzyme that inserts the DNA modification would disrupt the defense system, allowing the restriction enzyme to destroy the host cell’s DNA. In some bacteria, the modifications may also be essential as epigenetic marks and thus antibiotic targets as well.
“A lot of these enzymes are unique to the bacterium and they’re also essential to the survival of that organism, so they might make good antibiotic targets,” Dedon says.
Combination therapy may be better than radiotherapy alone to treat aggressive brain cancer
Posted by Acubiz | BlogJefferson researchers test approach that stops cancer cells from repairing themselves after radiotherapy
Radiotherapy effectively damages brain tumors but the cancer cells can repair themselves in order to live on. Now, researchers at Sidney Kimmel Cancer Center have tested a strategy that combines radiotherapy with a drug that shuts down the ability of tumor to mend themselves.
Researchers say their 12-patient study, published Jan. 29, 2016 online ahead of print in the Journal of Neuro-Oncology, offer enough promise that a more comprehensive, phase 2 clinical trial should be conducted to test the combination therapy for aggressive, recurrent brain cancer.
“We saw synergy between radiotherapy and the agent, panobinostat. Our findings suggest panobinostat makes radiotherapy much more effective,” says the study’s senior author, Yaacov R. Lawrence, M.D., of the Department of Radiation Oncology at Thomas Jefferson University’s Sidney Kimmel Medical College.
All 12 patients tested had high grade gliomas that had recurred after initial radiotherapy. Eight patients had recurrent glioblastoma, and four had recurrent anaplastic astrocytoma. These two forms of aggressive brain cancer represent almost 70 percent of newly diagnosed gliomas, which are diagnosed in about 10,000 patients annually. Despite response to initial radiation, most patients relapse within two years and overall survival is then limited to a year or less.
“There is no standard treatment for recurrent high grade gliomas. At Jefferson, we have a lot of experience with offering a second course of radiation after a patient relapses, in order to increase survival, but we are excited by the promise of a targeted agent that makes initial and repeat radiotherapy more effective,” says co-author Adam Dicker, M.D., Ph.D., FASTRO, Chair and Professor of Radiation Oncology, Pharmacology and Experimental Therapeutics at the Sidney Kimmel Medical College.
Panobinostat, approved for use in 2015 for treatment of multiple myeloma, is being tested in a variety of other cancers. It is a histone deacetylase inhibitor that has been shown to modify expression of about eight percent of RNA molecules produced from genes. Modifying RNA changes protein production, unsetting cancer growth. The drug also turns off RAD51, a DNA repair enzyme, Dr. Dicker says.
Researchers found that the highest dose of panobinostat tested in patients was well tolerated, and they observed improved progression-free survival and overall survival.
“The intent of this study was not to demonstrate benefit of the combination therapy, but to test safety. Still, we did note promising activity, which must be validated in further studies,” Dr. Lawrence says.
Female fertility is dependent on functional expression of the E3 ubiquitin ligase Itch
Posted by Acubiz | BlogThe post-translational addition of ubiquitin to proteins by enzymes of the E3 ubiquitin ligase family is largely recognized as a means to target misfolded or unwanted proteins for degradation by the proteasome. However, it is now understood that ubiquitination serves as a signal to modify a number of cellular functions such as protein trafficking, cell signaling, DNA repair, chromatin modifications, cell-cycle progression, and cell death. Though these functions are integral for all cells throughout the body, the physiologic role of specific E3 ligases must yet be defined in the context of various tissues. For example, very few studies exist that interrogate the function of specific E3 ubiquitin ligases in the reproductive system.
The physiologic roles of E3 ubiquitin ligases have been examined in knockout or mutant mouse models. In previous work with a mouse model that contained a loss of function mutation in the ITCH E3 ubiquitin ligase gene (mice termed itchy due to the chronic dermatitis phenotype) it was discovered that male mice displayed a number of alterations in testicular germ cells. Although there were phenotypic changes in the germ cells of the itchy male mice, fertility assays suggested that male reproduction remained functional. Itchy females, however, produced fewer offspring when bred to either itchy or wild type male mice. This led Richburg and colleagues from the University of Texas at Austin to evaluate the physiologic role of ITCH in the female reproductive system.
Their findings reported in the February 2016 issue of Experimental Biology and Medicine reveal several alterations in reproductive function in itchy female mice when compared to wild type female mice. Itchy females had both fewer implantations and tended to have fewer corpora lutea. Additionally, the itchy females remained in estrus longer, resulting in extended estrous cycles. The loss of ITCH within the ovary was confirmed, yet alterations in the expression of prototypical ITCH targets in the ovaries were not indicated. These results suggest the existence of an ovary-specific ITCH substrate or non-degradation dependent signaling pathway responsible for these phenotypic alterations. Alternatively, because ITCH works in the immune system to polarize T-cells towards an autoimmune type 2 activation state, these results may be indicative of immune interactions within the female reproductive system. The results of this work illustrate the functional participation of E3 ubiquitin ligases, specifically ITCH, in physiologic female reproduction. The lead author further reflects, “The female reproductive tract has long been recognized as a specialized immune environment, from macrophages that aid in luteal progression to T-cell tolerance in the uterus during fetal implantation. The results reported in this manuscript suggest that the Itchy mice could provide a useful model to evaluate the repercussions of preferential T-cell differentiation towards the type 2 phenotype on ovulation, estrus, and implantation.”
Researchers find association between oral bacteria and esophageal cancer
Posted by Acubiz | BlogFindings represent the first direct evidence that P. gingivalis could be a risk factor for esophageal cancer
University of Louisville School of Dentistry researchers have found a bacterial species responsible for gum disease, Porphyromonas gingivalis, is present in 61 percent of patients with esophageal squamous cell carcinoma (ESCC). The findings, published recently in Infectious Agents and Cancer, only detected P. gingivalis in 12 percent of tissues adjacent to the cancerous cells, while this organism was undetected in normal esophageal tissue.
“These findings provide the first direct evidence that P. gingivalis infection could be a novel risk factor for ESCC, and may also serve as a prognostic biomarker for this type of cancer,” said Huizhi Wang, M.D., Ph.D., assistant professor of oral immunology and infectious diseases at the UofL School of Dentistry. “These data, if confirmed, indicate that eradication of a common oral pathogen may contribute to a reduction in the significant number of people suffering with ESCC.”
The esophagus, a muscular tube critical to the movement of food from the mouth to the stomach, is lined with two main kinds of cells, thus there are two main types of esophageal cancer: adenocarcinoma and squamous cell carcinoma. The latter is more common in developing countries.
In collaboration with the College of Clinical Medicine of Henan University of Science and Technology in Luoyang, China, Wang and his UofL colleagues Richard J. Lamont, Ph.D., Jan Potempa, Ph.D., D.Sc., and David A. Scott, Ph.D., tested tissue samples from 100 patients with ESCC and 30 normal controls.
The research team measured the expression of lysine-gingipain, an enzyme unique to P. gingivalis, as well as the presence of the bacterial cell DNA within the esophageal tissues. Both the bacteria-distinguishing enzyme and its DNA were significantly higher in the cancerous tissue of ESCC patients than in surrounding tissue or normal control sites. The researchers also found the presence of P. gingivalis correlated with other factors, including cancer cell differentiation, metastasis and overall survival rate.
According to Wang, there are two likely explanations: either ESCC cells are a preferred niche for P. gingivalis to thrive or the infection of P. gingivalisfacilitates the development of esophageal cancer.
If the former is true, Wang says simple antibiotics may prove useful or researchers can develop other therapeutic approaches for esophageal cancer utilizing genetic technology to target the P. gingivalis and ultimately destroy the cancer cells.
“Should P. gingivalis prove to cause ESCC, the implications are enormous,” Wang said. “It would suggest that improving oral hygiene may reduce ESCC risk; screening for P. gingivalis in dental plaque may identify susceptible subjects; and using antibiotics or other anti-bacterial strategies may prevent ESCC progression.”
According to the Centers for Disease Control, about 15,000 people in the United States are diagnosed with esophageal cancer each year. As with most cancers, there are a number of risk factors including chemical exposure, diet, heredity and age. It is somewhat difficult to diagnosis this cancer early, and it is characterized by rapid progression and high mortality.