Potential treatment for Huntington’s disease, found effective, safe in mice, monkeys
Posted by Acubiz | BlogDrug enters clinical testing
A drug that would be the first to target the cause of Huntington’s disease (HD) is effective and safe when tested in mice and monkeys, according to data released today that will be presented at the American Academy of Neurology’s 68th Annual Meeting in Vancouver, Canada, April 15 to 21, 2016. A study to test the drug in humans has begun.
Huntington’s disease is a rare, hereditary disease that causes uncontrolled movements, loss of intellectual abilities, emotional problems and eventually death. The disease is passed from parent to child through a mutation in the huntingtin gene. The mutation results in the production of a disease-causing huntingtin protein. Each child has a 50/50 chance of inheriting the gene mutation. Everyone who inherits the mutated gene will eventually develop the disease.
The new drug, called IONIS-HTTRx, is an antisense drug that acts as a “gene silencer” to inhibit the production of huntingtin protein in people with Huntington’s disease.
“It is very exciting to have the possibility of a treatment that could alter the course of this devastating disease,” said clinical study principal investigator Blair R. Leavitt, MD, of the University of British Columbia in Vancouver. “Right now we only have treatments that work on the symptoms of the disease.” Leavitt notes the drug is still years away from being used in human clinical practice.
Earlier studies in mouse models of Huntington’s disease showed that treatment with antisense drugs delays disease progression and results in sustained reversal of the disease phenotype. In YAC128 mice, a transgenic model of HD, motor deficits improved within one month of initiating antisense treatment and were restored to normal at two months after treatment termination. Motor skills of antisense-treated BACHD mice, another transgenic model of HD, improved eight weeks after initiation of treatment and persisted for at least nine months after treatment termination. In monkeys, dose-dependent reductions in HTT mRNA and Htt protein throughout the central nervous system were observed after intrathecal administration of an antisense drug. Reduction of cortical huntingtin levels by 50 percent was readily achieved in monkeys and correlated with 15 to 20 percent reduction in the caudate. In further tests in rodents and monkeys, IONIS-HTTRx was found to be well-tolerated without any dose-limiting side effects.
The drug is now in a Phase 1/2a clinical study. The drug is delivered into the cerebral spinal fluid via an intrathecal injection in the lumbar space, as antisense drugs do not cross the blood brain barrier. In the current clinical study, the drug is administered in four doses at monthly intervals. Different doses of the drug will be evaluated for safety and tolerability. In addition, drug pharmacokinetics will be characterized and effects of the drug on specific biomarkers and clinical outcomes will be examined.
The drug was developed by scientists at Ionis Pharmaceuticals in collaboration with their partners CHDI Foundation, Roche Pharmaceuticals and academic collaborators at University of California, San Diego. The preclinical studies were conducted by Ionis Pharmaceuticals. The ongoing clinical study is supported by Ionis Pharmaceuticals and is part of Ionis’ collaboration with Roche to develop antisense drugs to treat Huntington’s disease.
Research on treatments for advanced ovarian cancer
Posted by Acubiz | BlogResearch led by a Dignity Health St. Joseph’s Hospital and Medical Center physician on ovarian cancer was published in the Feb. 24, 2016 issue ofNew England Journal of Medicine. The research was directed by Bradley J. Monk, M.D. and researchers at 12 other medical facilities around the nation.
The featured research titled “Every-3-Week vs. Weekly Paclitaxel and Carboplatin for Ovarian Cancer” unveils that the standard front-line treatment for advanced ovarian cancer should either be every three week chemotherapy with carboplatin, paclitaxel plus bevacizumab (an antibody than inhibits blood vessels from feeding the cancer) or every three week chemotherapy with carboplatin and weekly dose intense paclitaxel. The latter does not involve expensive bevacizumab known as Avastin but is more inconvenient.
“This supports the use of weekly chemotherapy without bevacizumab in treating advanced ovarian cancer,” says Dr. Monk who is Director, Division of Gynecologic Oncology Vice Chair, Department of Obstetrics and Gynecology University of Arizona Cancer Center at St. Joseph’s Hospital and senior author of the publication.
The research was funded by NRG Oncology (formerly the Gynecologic Oncocolgy Group) which is part of the National Institutes of Health and enrolled patients at cancer centers around the country. Many women with ovarian cancer at the University of Arizona Cancer Center at St. Joseph’s Hospital participated in this study.
Importantly, this study did not investigate intraperitoneal chemotherapy (IP) where the chemotherapy is infused directly into the belly. Many believe that the regimen of dose intense weekly chemotherapy supported by the current publication capitalizes on the same key components of IP chemotherapy namely higher doses and weekly administration. However, the intravenous recipe supported by this publication does did not show the intense side effects seen with IP treatments.
This advance is only a small step forward as newer appraises to ovarian cancer are being developed at University of Arizona Cancer Center at St. Joseph’s Hospital. For example, doctors are studying a unique class of drugs called PARP inhibitors as well as immunotherapy. The latter uses recently discovered approaches to re-programming a woman’s immune system to recognize and fight her ovarian cancer. Such drugs have recently been FDA approved to treat lung cancer and melanoma.
Breast reconstruction using abdominal tissue: Differences in outcome with four different techniques
Posted by Acubiz | BlogIn women undergoing breast reconstruction using their own (autologous) tissue, newer “muscle-sparing” abdominal flaps can reduce complications while improving some aspects of quality of life, reports a study in the March issue of Plastic and Reconstructive Surgery®, the official medical journal of the American Society of Plastic Surgeons (ASPS).
Comparison of four types of abdominal flaps used for autologous breast shows reconstruction shows differences in some key outcomes–notably related to problems related to hernias or bulging at the abdominal “donor site,” according to the new research by Dr. Sheina A. Macadam of University of British Columbia, Vancouver, and colleagues.
Four Abdominal Flaps Show Differences in Satisfaction and Other Key Outcomes
The researchers identified nearly 1,800 women undergoing autologous breast reconstruction after mastectomy at five US and Canadian university hospitals. The reconstructions were done using different types of abdominal flaps:
• Traditional flaps incorporating the rectus muscle of the abdomen–called the free transverse abdominis myocutaneous flap (f TRAM) or pedicled transverse abdominis myocutaneous flap (p TRAM)
• Newer flaps that avoid or use only a part of the rectus muscle–called the deep inferior epigastric artery perforator flap (DIEP) or muscle-sparing free transverse abdominis myocutaneous flap (msf TRAM)
Hospital records were used to compare complication rates across the four flap types. In addition, about half of the women completed the BREAST-Q© questionnaire, which assesses various aspects of quality of life after breast reconstruction. Average follow-up time was 5.5 years.
The most serious types of complications–including total flap loss and abnormal blood clots–were not significantly different between groups. Both the DIEP and msf-TRAM flaps were associated with lower rates of fat necrosis, compared to the p-TRAM flap.
The p-TRAM flap was also associated with the highest rate of abdominal hernia or bulging: nearly 17 percent. The risk of these donor-site complications was about eight percent with the msf-TRAM flap, six percent with the f-TRAM flap, and four percent with the DIEP flap.
That was consistent with the BREAST-Q responses, which showed better scores on a subscale reflecting physical issues related to the abdomen with the DIEP flap compared to the p-TRAM flap. Scores on the other BREAST-Q subscales–reflecting overall satisfaction with the breasts, and psychosocial and sexual well-being–were similar across groups.
About 20 percent of women undergoing breast reconstruction choose autologous reconstruction, which is most often done using abdominal flaps. Muscle-sparing flaps such as the DIEP and msf-TRAM have been developed to reduce complications at the abdominal donor site. While more women are interested in these muscle-sparing approaches, it hasn’t been clear whether the longer operative times and increased costs of these procedures are justified by improved outcomes.
The new study shows some differences in complications and patient-reported outcomes with different abdominal flaps. “The DIEP was associated with the highest abdominal well-being and the lowest abdominal morbidity when compared to the p-TRAM, but did not differ from msf-TRAM and f-TRAM,” Dr. Macadam and coauthors write.
While formal randomized trials would be needed to confirm these results, the researchers believe their findings provide important evidence on the outcomes of current options for autologous breast reconstruction. They conclude, “The differences we have found in patient-reported symptoms and abdominal donor site outcomes may shift the practice of plastic surgeons towards utilizing methods with lower donor site [complications] and higher patient-reported satisfaction.”
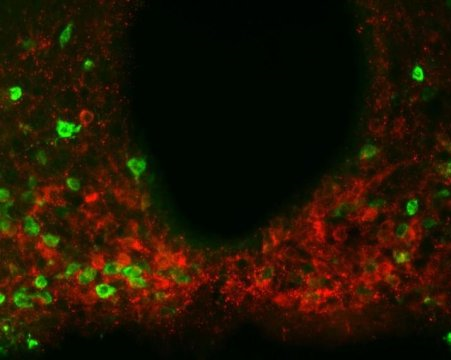
Metabolism protein found to also regulate feeding behavior in the brain
Posted by Acubiz | BlogScientists found evidence of the metabolism-regulating protein amylin, shown in red, present in multiple regions throughout a brain area called the hypothalamus. Experiments suggest amylin produced by hypothalamic neurons helps reduce food consumption together with leptin.
Credit: Laboratory of Molecular Genetics at The Rockefeller University/Cell Metabolism
The molecular intricacies of hunger and satiety, pivotal for understanding metabolic disorders and the problem of obesity, are not yet fully understood by scientists. However, new research from The Rockefeller University reveals an important new component of the system responsible for regulating food intake: a hormone called amylin, which acts in the brain to help control consumption.
“How much a person eats is regulated by a complex circuit, and in order to understand it, we need to identify all the molecules involved,” says Jeffrey Friedman, Marilyn M. Simpson Professor and head of the Laboratory of Molecular Genetics at Rockefeller. “Amylin caught our attention when we were profiling a set of neurons in the hypothalamus, a part of the brain known to be involved in feeding behavior. Because it plays a role in sugar metabolism elsewhere in the body, we were interested in exploring its function in the brain.”
What stops us from eating too much?
Friedman is well known for his 1994 discovery of the hormone leptin, one regulator in this process. Defects in leptin production are associated with obesity. However, treating obesity with leptin alone has not proven effective except in cases of severe leptin deficiency, suggesting that additional components are involved in this system.
The findings, published recently in Cell Metabolism, suggest that leptin and amylin work in concert to control food intake and body weight.
Friedman and colleagues first identified the precursor to amylin — called Iselt amyloid peptide (Iapp) — in the brain by using a technology known as translating ribosome affinity purification, previously developed by fellow Rockefeller scientists. The researchers found that Iapp is abundant in multiple regions throughout the hypothalamus. (Incidentally, these findings contradict previous results, suggesting that prior experiments, which have not consistently found amylin in the brain, may not have used techniques sensitive enough to detect Iapp.)
To tease out the function of amylin in the hypothalamus, the researchers assessed its presence in mice that were obese due to leptin deficiency. When these mice were given leptin, their Iapp levels increased significantly, indicating that leptin regulates the expression of amylin.
Molecular teamwork
“We also looked directly at how amylin and leptin affect feeding behavior,” says lead author Zhiying Li, a research associate in the lab. “When we give leptin to mice, it significantly suppresses food intake. However, when we give leptin to mice in which amylin is rendered nonfunctional with an inhibitor, the effect of leptin is blunted. This means that leptin and amylin are working together in a way that reduces feeding.”
Additionally, the researchers evaluated how amylin controls neural signals. From recordings of neuron signals showing that leptin and amylin act on the same neurons in similar ways, they hypothesize that these hormones act in a synergistic manner, working together to produce an enhanced neural signal.
“These findings confirm a functional role for amylin in the central nervous system, and provide a potential mechanism to treat obesity more effectively, through combination therapy,” says Friedman. “While this is a piece of the puzzle, we still need a better understanding of the cellular mechanisms involved in this system, which could provide new approaches that involve improved leptin signaling and sensitivity.”
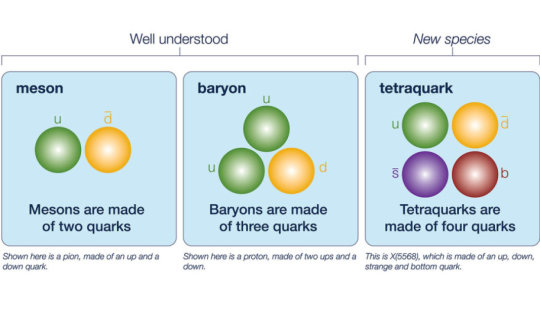
Tetraquarks: New four-flavor particle discovered
Posted by Acubiz | BlogScientists have discovered a new particle — the latest member to be added to the exotic species of particle known as tetraquarks.
Credit: Artwork by Fermilab
Scientists on the DZero collaboration at the U.S. Department of Energy’s Fermilab have discovered a new particle — the latest member to be added to the exotic species of particle known as tetraquarks.
Quarks are point-like particles that typically come in packages of two or three, the most familiar of which are the proton and neutron (each is made of three quarks). There are six types, or “flavors,” of quark to choose from: up, down, strange, charm, bottom and top. Each of these also has an antimatter counterpart.
Over the last 60 years, scientists have observed hundreds of combinations of quark duos and trios.
In 2008 scientists on the Belle experiment in Japan reported the first evidence of quarks hanging out as a foursome, forming a tetraquark. Since then physicists have glimpsed a handful of different tetraquark candidates, including now the recent discovery by DZero — the first observed to contain four different quark flavors.
DZero is one of two experiments at Fermilab’s Tevatron collider. Although the Tevatron was retired in 2011, the experiments continue to analyze billions of previously recorded events from its collisions.
As is the case with many discoveries, the tetraquark observation came as a surprise when DZero scientists first saw hints in July 2015 of the new particle, called X(5568), named for its mass — 5568 megaelectronvolts.
“At first, we didn’t believe it was a new particle,” says DZero co-spokesperson Dmitri Denisov. “Only after we performed multiple cross-checks did we start to believe that the signal we saw could not be explained by backgrounds or known processes, but was evidence of a new particle.”
And the X(5568) is not just any new tetraquark. While all other observed tetraquarks contain at least two of the same flavor, X(5568) has four different flavors: up, down, strange and bottom.
“The next question will be to understand how the four quarks are put together,” says DZero co-spokesperson Paul Grannis. “They could all be scrunched together in one tight ball, or they might be one pair of tightly bound quarks that revolves at some distance from the other pair.”
Four-quark states are rare, and although there’s nothing in nature that forbids the formation of a tetraquark, scientists don’t understand them nearly as well as they do two- and three-quark states.
This latest discovery comes on the heels of the first observation of a pentaquark — a five-quark particle — announced last year by the LHCb experiment at the Large Hadron Collider.
Scientists will sharpen their picture of the quark quartet by making measurements of properties such as the ways X(5568) decays or how much it spins on its axis. Like investigations of the tetraquarks that came before it, the studies of the X(5568) will provide another window into the workings of the strong force that holds these particles together.
And perhaps the emerging tetraquark species will become an established class in the future, showing themselves to be as numerous as their two- and three-quark siblings.
“The discovery of a unique member of the tetraquark family with four different quark flavors will help theorists develop models that will allow for a deeper understanding of these particles,” says Fermilab Director Nigel Lockyer.
Seventy-five institutions from 18 countries collaborated on this result from DZero.
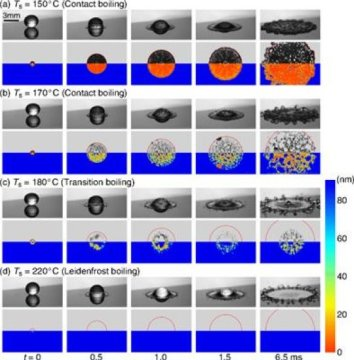
First images of the nanolayer beneath a dancing Leidenfrost droplet
Posted by Acubiz | BlogCredit: Image courtesy of University of Twente
Water droplets on a very hot plate don’t evaporate but levitate and move around: this is known as the Leidenfrost effect and it always guarantees beautiful images. For the first time, researchers of the Physics of Fluids group of the University of Twente (MESA+ Institute for Nanotechnology) have made images of the tiny layer beneath the droplet, when it impacts on the surface. Thanks to this images, a more detailed explanation of the phenomenon is possible, the researchers conclude in their publication in Physical Review Letters.
A droplet of water impacting on a plate that is at the water boiling point, will spread and evaporate fast. On a surface that is much hotter, something else happens: the droplet will levitate on top of its own vapor. For this phenomenon to occur, the temperature must have reached the Leidenfrost value. On impact, the levitation is approx. 100 nanometer (a nanometer is one millionth of a millimeter). The UT scientist now found a way to get detailed images of this. “We combine high-speed images with a laser technique called total internal reflection imaging,” PhD student Michiel van Limbeek explains. “Using this laser we can distinguish wet and dry areas: does the liquid touch the surface or, in case of Leidenfrost, doesn’t it at all?”
In this case, ethanol was chosen instead of water. The droplets fall onto a sapphire surface. The researchers distinguish three boiling regimes: contact boiling, transition boiling and Leidenfrost boiling. The TIR-measurements show the wet and dry spots on the surface, thanks to the different refractive index of vapor and ethanol: the laser light is only reflected when it meets vapor. Thanks to the TIR images, more information is obtained about the incident angle and the spreading radius: the different regimes can be distinguished better. In the transition region towards Leidenfrost, for example, the center part of the drop still contacts the surface (wetting) while the outer ring already levitates.
Neck and dimple
A remarkable conclusion is that the droplet is not flat at the bottom, as is often assumed in ‘pancake models’ describing Leidenfrost. The images show the shape of a ring: to be more precise, a combination of a ‘neck’ at the closest distance to the surface and a ‘dimple’ which is further away from the surface towards the center of the drop. This results in a better quantitative description.
The new experiments and theory make it possible to describe the Leidenfrost effect for a wide range of fluids, surfaces and temperature regimes. In many applications where there is heat transfer from solids to liquids this is of great value, like in cooling techniques, engines or more efficient chemical reactors.
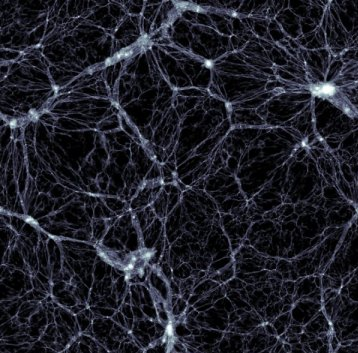
Black holes banish matter into cosmic voids
Posted by Acubiz | BlogDistribution of dark matter, with a width and height of 350 million light-years and a thickness of 300000 light years. Galaxies are found in the small, white, high-density dots.
Credit: Markus Haider / Illustris collaboration.
We live in a universe dominated by unseen matter, and on the largest scales, galaxies and everything they contain are concentrated into filaments that stretch around the edge of enormous voids. Thought to be almost empty until now, a group of astronomers based in Austria, Germany and the United States now believe these dark holes could contain as much as 20% of the ‘normal’ matter in the cosmos and that galaxies make up only 1/500th of the volume of the universe. The team, led by Dr Markus Haider of the Institute of Astro- and Particle Physics at the University of Innsbruck in Austria, publish their results in a new paper in Monthly Notices of the Royal Astronomical Society.
Looking at cosmic microwave radiation, modern satellite observatories like COBE, WMAP and Planck have gradually refined our understanding of the composition of the universe, and the most recent measurements suggest it consists of 4.9% ‘normal’ matter (i.e. the matter that makes up stars, planets, gas and dust), or ‘baryons’, whereas 26.8% is the mysterious and unseen ‘dark’ matter and 68.3% is the even more mysterious ‘dark energy’.
Complementing these missions, ground-based observatories have mapped the positions of galaxies and, indirectly, their associated dark matter over large volumes, showing that they are located in filaments that make up a ‘cosmic web’. Haider and his team investigated this in more detail, using data from the Illustris project, a large computer simulation of the evolution and formation of galaxies, to measure the mass and volume of these filaments and the galaxies within them.
Illustris simulates a cube of space in the universe, measuring some 350 million light years on each side. It starts when the universe was just 12 million years old, a small fraction of its current age, and tracks how gravity and the flow of matter changes the structure of the cosmos up to the present day. The simulation deals with both normal and dark matter, with the most important effect being the gravitational pull of the dark matter.
When the scientists looked at the data, they found that about 50% of the total mass of the universe is in the places where galaxies reside, compressed into a volume of 0.2% of the universe we see, and a further 44% is in the enveloping filaments. Just 6% is located in the voids, which make up 80% of the volume.
But Haider’s team also found that a surprising fraction of normal matter — 20% — is likely to be have been transported into the voids. The culprit appears to be the supermassive black holes found in the centres of galaxies. Some of the matter falling towards the holes is converted into energy. This energy is delivered to the surrounding gas, and leads to large outflows of matter, which stretch for hundreds of thousands of light years from the black holes, reaching far beyond the extent of their host galaxies.
Apart from filling the voids with more matter than thought, the result might help explain the missing baryon problem, where astronomers do not see the amount of normal matter predicted by their models.
Dr Haider comments: “This simulation, one of the most sophisticated ever run, suggests that the black holes at the centre of every galaxy are helping to send matter into the loneliest places in the universe. What we want to do now is refine our model, and confirm these initial findings.”
Illustris is now running new simulations, and results from these should be available in a few months, with the researchers keen to see whether for example their understanding of black hole output is right. Whatever the outcome, it will be hard to see the matter in the voids, as this is likely to be very tenuous, and too cool to emit the X-rays that would make it detectable by satellites.
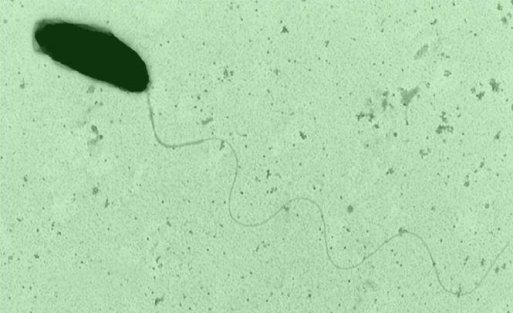
Bacteria take ‘RNA mug shots’ of threatening viruses
Posted by Acubiz | BlogElectron micrograph of the marine bacteria Marinomonas mediterranea is shown.
Credit: Antonio Sanchez-Amat
Scientists from The University of Texas at Austin, the Stanford University School of Medicine and two other institutions have discovered that bacteria have a system that can recognize and disrupt dangerous viruses using a newly identified mechanism involving ribonucleic acid (RNA). It is similar to the CRISPR/Cas system that captures foreign DNA. The discovery might lead to better ways to thwart viruses that kill agricultural crops and interfere with the production of dairy products such as cheese and yogurt.
The research appears online Feb. 25 in the journal Science.
Both RNA and DNA are critical for life. In humans and many other organisms, DNA molecules act as the body’s blueprints, while RNA molecules act as the construction crew–reading the blueprints, building the body and maintaining the functions of life.
The research team found for the first time that bacteria can snatch bits of RNA from invaders such as viruses and incorporate the RNA into their own genomes, using this information as something akin to mug shots. They then help the bacteria recognize and disrupt dangerous viruses in the future.
“This mechanism serves a defensive purpose in bacteria,” says Alan Lambowitz, director of the Institute for Cellular and Molecular Biology at UT Austin and co-senior author of the paper. “You could imagine transplanting it into other organisms and using it as a kind of virus detector.”
The newly discovered mechanism stores both DNA and RNA mug shots from viruses in a bacterium’s genome. That makes sense from an evolutionary standpoint, the researchers say, given that some viruses are DNA-based and some are RNA-based.
Lambowitz says that as a next step, researchers can examine how to genetically engineer a crop such as tomatoes so that each of their cells would carry this virus detector. Then the researchers could do controlled laboratory experiments in which they alter environmental conditions to see what effects the changes have on the transmission of pathogens.
“Combining these plants with the environment that they face, be it natural or involving the application of herbicides, insecticides or fungicides, could lead to the discovery of how pathogens are getting to these plants and what potential vectors could be,” says Georg Mohr, a research associate at UT Austin and co-first author of the paper.
Another application might be in the dairy industry, where viruses routinely infect the bacteria that produce cheese and yogurt, causing the production process to slow down or even preventing it from going to completion. Currently, preventing infections is complicated and costly. Lambowitz and Mohr say dairy bacteria could be engineered to record their virus interactions and defend against subsequent infections.
This RNA-based defense mechanism is closely related to a previously discovered mechanism, called CRISPR/Cas, in which bacteria snatch bits of DNA and store them as mug shots. That method has inspired a new way of editing the genomes of virtually any living organism, launching a revolution in biological research and sparking a patent war, but the researchers say they do not anticipate this new discovery will play a role in that sort of gene-editing. However, the enzymatic mechanism used to incorporate RNA segments into the genome is novel and has potential biotechnological applications.
Researchers discovered this novel defense mechanism in a type of bacteria commonly found in the ocean called Marinomonas mediterranea. It’s part of a class of microbes called Gammaproteobacteria, which include many human pathogens such as those that cause cholera, plague, lung infections and food poisoning.

Fungi are at the root of tropical forest diversity, or lack thereof, study finds
Posted by Acubiz | BlogThe study focused on tropical forest patches dominated by one tree species, Oreomunnea mexicana, pictured.
Credit: Photo: James Dalling
The types of beneficial fungi that associate with tree roots can alter the fate of a patch of tropical forest, boosting plant diversity or, conversely, giving one tree species a distinct advantage over many others, researchers report.
Their study, reported in the journal Ecology Letters, sought to explain a baffling phenomenon in some tropical forests: Small patches of “monodominant forest,” where one species makes up more than 60 percent of the trees, form islands of low diversity in the otherwise highly diverse tropical forest growing all around them.
The new study focused on mountain forests in Panama that harbor hundreds of tree species, but which include small patches dominated by the tree species Oreomunnea mexicana.
“Tropical ecologists are puzzled by how so many species co-occur in a tropical forest,” said University of Illinois plant biology professor James Dalling, who led the study with graduate student Adriana Corrales and collaborators from Washington University in St. Louis and the Smithsonian Tropical Research Institute in Panama. “If one tree species is a slightly better competitor in a particular environment, you would expect its population to increase and gradually exclude other species.”
That doesn’t happen often in tropical forests, however, he said. Diversity remains high, and patches dominated by a single species are rare. Understanding how monodominant forests arise and persist could help explain how tropical forests otherwise maintain their remarkable diversity, he said.
The researchers focused on two types of fungi that form symbiotic relationships with trees: arbuscular mycorrhizas and ectomycorrhizas. Arbuscular mycorrhizas grow inside the roots of many different tree species, supplying phosphorus to their tree hosts. Ectomycorrhizas grow on the surface of tree roots and draw nitrogen from the soil, some of which they exchange for sugars from the trees. Ectomycorrhizas cooperate with only a few tree species — 6 percent or less of those that grow in tropical forests.
Previous studies found that arbuscular mycorrhizas commonly occur in the most diverse tropical forests, while ectomycorrhizal fungi dominate low-diversity patches.
“When you walk in a patch of forest where 70 percent of the trees belong to a single species that also happens to be an ectomycorrhizal-associated tree, it makes you think there is something going on with the fungi that could be mediating the formation of these monodominant forests,” Corrales said.
The researchers tested three hypotheses to explain the high abundance ofOreomunnea. First, they tested the idea that Oreomunnea trees are better able to resist species-specific pathogens than trees growing in more diverse forest areas.
“We were expecting that Oreomunnea seedlings would grow better in soil coming from beneath other Oreomunnea trees, because that’s how the tree grows in nature,” Corrales said. “But we found the opposite: TheOreomunnea suffered more from pathogen infection when grown in soil from the same species than in soil from other species.”
The researchers next tested whether mature Oreomunnea trees supported nearby Oreomunnea seedlings by sending sugars to them via a shared network of ectomycorrhizal fungi. But they found no evidence of cooperation between the trees.
“The seedlings that were isolated from the ectomycorrhizas of otherOreomunnea trees grew better than those that were in contact with the fungi from other trees of the same species,” Corrales said.
In a third set of experiments, the team looked at the availability of nitrogen inside and outside the Oreomunnea patches.
“We saw that inorganic nitrogen was much higher outside than inside the patches,” Corrales said. Tree species that normally grow outside the patches did well on the high-nitrogen soils, but suffered when transplanted inside theOreomunnea patches. A look at the nitrogen isotopes in the fungi, soils and in the seedlings’ leaves revealed the underlying mechanism by which the fungi influenced the species growing inside and outside the Oreomunneapatches.
The team found evidence consistent with ectomycorrhizal uptake of nitrogen directly from decomposing material in the soil. These fungi make some of their nitrogen available to the Oreomunnea trees while starving other plants and soil microbes of this essential nutrient, Corrales said. The lack of adequate nitrogen means bacteria and fungi are unable to break down organic matter in the soil, causing most other trees to suffer because they depend on the nitrogen supplied by microbial decomposers, she said.
“We found a novel mechanism that can explain why certain tree species in tropical forests are highly abundant, and that is because their fungi provide them with a source of nitrogen that is not accessible to competing species,” Dalling said. “So they have an advantage because their competitors are now starved of nitrogen.”
Researchers have found recently that similar processes can occur in temperate forests, but this is the first study to link this process to tropical forest monodominance, Dalling said.
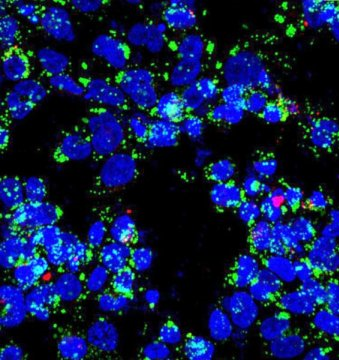
New theorem helps reveal tuberculosis’ secret
Posted by Acubiz | BlogUncovering missing connections in biochemical networks
Upon infection with Mycobacterium tuberculosis bacilli (labeled in red), macrophages (nuclei stained blue) accumulate lipid droplets (green). The network controlling the expression of an enzyme that is central to bacterial metabolic switching to lipids as nutrients during infection is the topic of a new paper by researchers at Rice and Rutgers universities.
Credit: Emma Rey-Jurado/Public Health Research Institute
A new methodology developed by researchers at Rice and Rutgers universities could help scientists understand how and why a biochemical network doesn’t always perform as expected. To test the approach, they analyzed the stress response of bacteria that cause tuberculosis and predicted novel interactions.
The results are described in a PLOS Computational Biology paper published today.
“Over the last several decades, bioscientists have generated a vast amount of information on biochemical networks, a collection of reactions that occur inside living cells,” said principal investigator Oleg Igoshin, a Rice associate professor of bioengineering.
“We are beginning to understand how these networks control the dynamics of a biological response, that is, the precise nature of how a concentration of biomolecules changes with time,” he said. “But to date, only a few general rules that relate the dynamical responses with the structure of the underlying networks have been formulated. Our theorem provides another such rule and therefore can be widely applicable.”
The theorem uses approaches from control theory, an interdisciplinary branch of engineering and mathematics that deals with the behavior of dynamical systems that have inputs. The theorem formulates a condition for an underlying biochemical network to display non-monotonic dynamics in response to a monotonic trigger. For instance, it would explain the expression of a gene that first speeds up, then slows down and returns to normal. (Monotonic responses always increase or always decrease; non-monotonic responses increase and then decrease, or vice-versa.)
The theorem states that a non-monotonic response is only possible if the system’s output receives conflicting messages from the input, such that one branch of the pathway activates it and another one deactivates it.
If a non-monotonic response is observed in a system that appears to be missing such conflicting paths, it would imply that some biochemical interactions remain undiscovered, Igoshin said.
“What we do is figure out the mechanism for a dynamic phenomenon that people have observed but can’t explain and that seems to be inconsistent with the current state of knowledge,” he said.
The theorem was formulated and proven in collaboration with Eduardo Sontag, a distinguished professor in the Department of Mathematics and Center for Quantitative Biology at Rutgers. Sontag focuses on general principles derived from feedback control analysis of cell signaling pathways and genetic networks.
The researchers applied their theory to explain how Mycobacterium tuberculosis responds to stresses that mimic those the immune system uses to fight the pathogen. Igoshin said M. tuberculosis is a master in surviving such stresses. Instead of dying, they become dormant Trojan horses that future conditions may reactivate.
According to the World Health Organization, a third of the world’s population is infected by the tuberculosis bacteria, though the disease kills only a fraction of those infected.
“The good thing is that 95 percent of infected people don’t have symptoms,” said Joao Ascensao, a Rice senior majoring in bioengineering and first author of the paper. “The bad thing is you can’t kill the bacteria. And then if you get immunodeficiency, due to HIV, starvation or other things, you’re out of luck because the disease will reactivate.”
Ascensao said M. tuberculosis is hard to grow and work with in a molecular biology setting. “A generation of E. coli takes 20 minutes to grow, but for M. tuberculosis, a generation takes from 24 hours to over 100 hours when it goes latent,” he said. “So even though we have this really sparse data, the theory allowed us to uncover what’s happening behind the scenes.”
The study was motivated by a 2010 publication by Marila Gennaro, a professor of Medicine in the Public Health Research Institute at Rutgers, and Pratik Datta, a research scientist in her lab, who are also co-authors of the new paper. Their results showed that as M. tuberculosis gradually runs out of oxygen, the expression of some genes would suddenly rise and then fall back. They characterized the biochemical network that controls the expression of these non-monotonic genes, but the mechanism of the dynamical response was not understood.
“It didn’t make sense to me intuitively,” Igoshin said. “At first I couldn’t prove it mathematically, but then Sontag’s theorem allowed us to conclude that some biochemical interactions were missing in the underlying network.”
Ascensao and Baris Hancioglu, then a postdoc in Igoshin’s lab and now a bioinformatics specialist at Ohio State University, built computer models and ran simulations of oxygen-starved M. tuberculosis. Their results suggested a few possible solutions that were tested in the follow-up experiments by Gennaro’s group.
Eventually the simulations predicted a new interaction that could explain the dynamics of the glyoxylate shunt genes that control the metabolic transition network known to be important to the bacteria’s virulence.
“Researchers found that the hypoxic (oxygen-starved) signal would lead bacteria to switch from one type of food to a different type of food,” Igoshin said. “They used to eat sugars, but they’d start eating the fat accumulated inside of infected macrophages, a type of immune cell. It looks like this switch might be associated with going from an active bacterium to a latent, dormant bacterium that’s stable and doesn’t cause any symptoms.”
The researchers argued that the stress-induced activation of adaptive metabolic pathways involving glyoxylate genes is transient, increasing only until there’s enough of the protein present to achieve stability. “If these hypotheses are correct,” they wrote, “drugs blocking negative interactions responsible for non-monotonic dynamics could in principle destabilize transitions to latency or trigger reactivation.”
The research was supported by the National Institutes of Health. The researchers used the National Science Foundation-supported supercomputing resources administered by Rice’s Ken Kennedy Institute for Information Technology.